 ORTEP-III
ORTEP-III

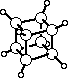
The standard example for illustrating the use of ORTEP is the novel compound cubane
(C8H8),
whose carbon-carbon bonds lie along the edges of a cube within experimental error.
The structure was published in 1964 by E. B. Fleischer (Journal of the American
Chemical Society, Vol. 86, p.3889).
The compound crystallizes with the trigonal symmetry of space group
R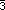 . The
. The 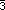 axis lies
along a body diagonal of the molecule, and as a result the compound contains only four
unique atoms. These are one carbon and its attached hydrogen in
general positions off
the
axis lies
along a body diagonal of the molecule, and as a result the compound contains only four
unique atoms. These are one carbon and its attached hydrogen in
general positions off
the 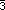 axis (C1 and H1) and one carbon and its hydrogen in special positions on
the
axis (C1 and H1) and one carbon and its hydrogen in special positions on
the 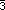 axis (C2 and
H2). Anisotropic temperature factor coefficients were fitted to the carbon atoms during the
least-squares refinement of the structure, and isotropic temperature factors were used for the
hydrogen atoms. The anisotropic temperature factors given for the carbon atoms are of the
type called 0 in ORTEP.
axis (C2 and
H2). Anisotropic temperature factor coefficients were fitted to the carbon atoms during the
least-squares refinement of the structure, and isotropic temperature factors were used for the
hydrogen atoms. The anisotropic temperature factors given for the carbon atoms are of the
type called 0 in ORTEP.
To draw the structure the following information is needed:
 =
=
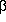 =
=
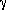 = 72.26o
= 72.26o
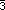 :
:
Positional Parameters (x,y,z in fractional parts along unit cell edges):
Anisotropic Temperature Factor Coefficients (b11, b22, b33, b12, b13, b23)
CUBANE E.B.FLEISCHER 1964 J.A.C.S. 86, 3889
1 5.34 5.34 5.34 72.26 72.26 72.26
x, y, z z, x, y y, z, x -x, -y, -z -z, -x, -y 1 -y, -z, -x
Atoms 1 and 2 are entered with positional parameter type 0 and anisotropic temperature factor type 0.
C1 -.18711 .19519 .10706 0 0 .04100 .04250 .04500 -.00420 -.01420 -.00510 0 C2 .11546 .11546 .11546 0 0 .04680 .04680 .04680 -.01430 -.01430 -.01430 0Atoms 3 and 4 are entered with positional parameter type 0 and with dummy sphere temperature factors (type 7) with a radius of 0.1 Å before scaling.
H1 -.32460 .34680 .18480 0 0 .10 7 H2 .21000 .21000 .21000 0 0 .10 7A dummy atom (atom 5) at the cell origin is also included with a blank card dummy sphere. This could also be entered with type 7 as were atoms 3 and 4.
ORGN 0 0 0 0 1 0
A 101 instruction is used to obtain a tabulation of the atoms surrounding one atom or a series of several designated atoms. For example, to obtain a list of all atoms (hydrogen and carbon atoms) out to a distance of 3.61 Å about the two carbons C1 and C2, the following 101 instruction would be used.
101 155501 2 1 4 3.61
|_______________| |__________| |____|
(a) (b) (c)
where the parts designate:
(b) target atoms 1 through 4 of all symmetry and translation operations
(c) a distance Dmax of 3.61 Å
A 102 instruction gives both interatomic distances and interatomic angles. The following instruction could be used to find all covalent bonds and bond angles about the two carbons.
102 155501 2 1 4 1.8
In this case a smaller Dmax
was used so that only the distances and
angles of immediate interest would be computed since there are n(n - 1)/2
angles for n interatomic vectors about an atom.
201
 5 inch boundary is specified with a 0.5 inch margin to
give a 4
5 inch boundary is specified with a 0.5 inch margin to
give a 4
 4 inch square working area. A
15 inch view distance might be reasonable to use
in viewing a model of this size. These are set with the
301 instruction.
4 inch square working area. A
15 inch view distance might be reasonable to use
in viewing a model of this size. These are set with the
301 instruction.
301 5.0 5.0 15. 0.5
402 555501 5 1 4 3.2
|_______________| |__________| |___|
(a) (b) (c)
where the parts designate:
(b) a run of target atoms from atom 1 to atom 4
(c) a sphere radius of 3.2 Å
501 555501 255501 155501 255501 155502 0 0
502 2 25. 1 28.
604 -50
503 2 2.7
The 1101 instruction starts the save sequence.
1101
2 1001
1 4 1 4 4 0.9 1.6 .04
1 716 .12 .30 .40
1 2
The 716 instruction is used, rather than 706, to shorten the output listing of the
example. A 706 instruction would provide a listing of all coordinates.
The hydrogen atoms (ANR = 3, 4) are to be drawn with a different standard model (instruction 702/712) than the carbon atoms. Chemical symbols are 0.12 in. high and offset 0.25 in. horizontally and vertically.
1 712 .12 .25 .25
3 4
2 812
1 4 1 4 4 0.9 1.6 .04
|____||____||_| |_________| |___|
(a) (b) (c) (d) (e)
where the parts designate:
(b) target atom number run of atoms making up second member of bond atom pairs
(c) bond type 4
(d) the distance range 0.9 to 1.6 Å that will cover all covalent bond distances
(e) the bond radius is 0.04 Å
The remaining fields on the card can be blank, since a complete set of bond distance labels is not desired.
906 155504 155503 .12 0 -.4
916 255504 155505 .12 0 -.4
Finally, a caption for the illustration is drawn. This can conveniently be positioned by
"hanging" it from the dummy atom 555501 and "bouncing" it 3 in. from the left x
boundary and 0.5 in. from the lower y boundary. The caption is 0.2 in. high.
3 902 555501 3.0 0.5 .20 0 0 Cubane
1102
202 4.0
503 2 -2.7
1103
202
-1
Page last revised: